Metagenomics with The Banfield Lab
The Banfield Lab
Background
It’s easy to think that we’ve discovered most of the species on the planet. In fact, the booming field of metagenomics – the study of genomic sequences – is using big data to help scientists better understand new and vast unexplored regions of the natural world: the microbiome. In the last few years, the price of genetic sequencing has plummeted to the point where scientists can now study ecosystems in their entirety, not just the parts of it that they already know how to identify. The Banfield Lab at UC Berkeley asked Stamen to help them bring this data to life.
What we built
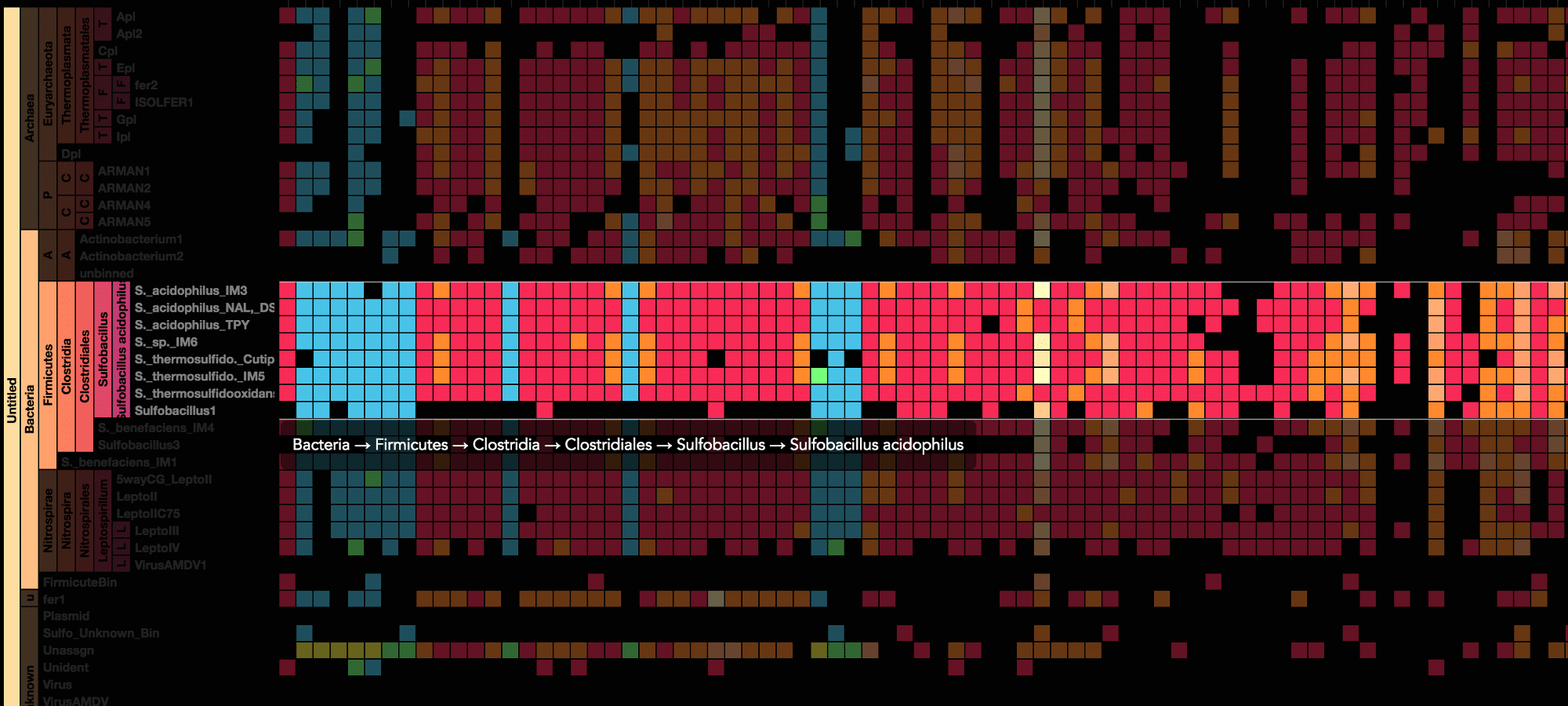
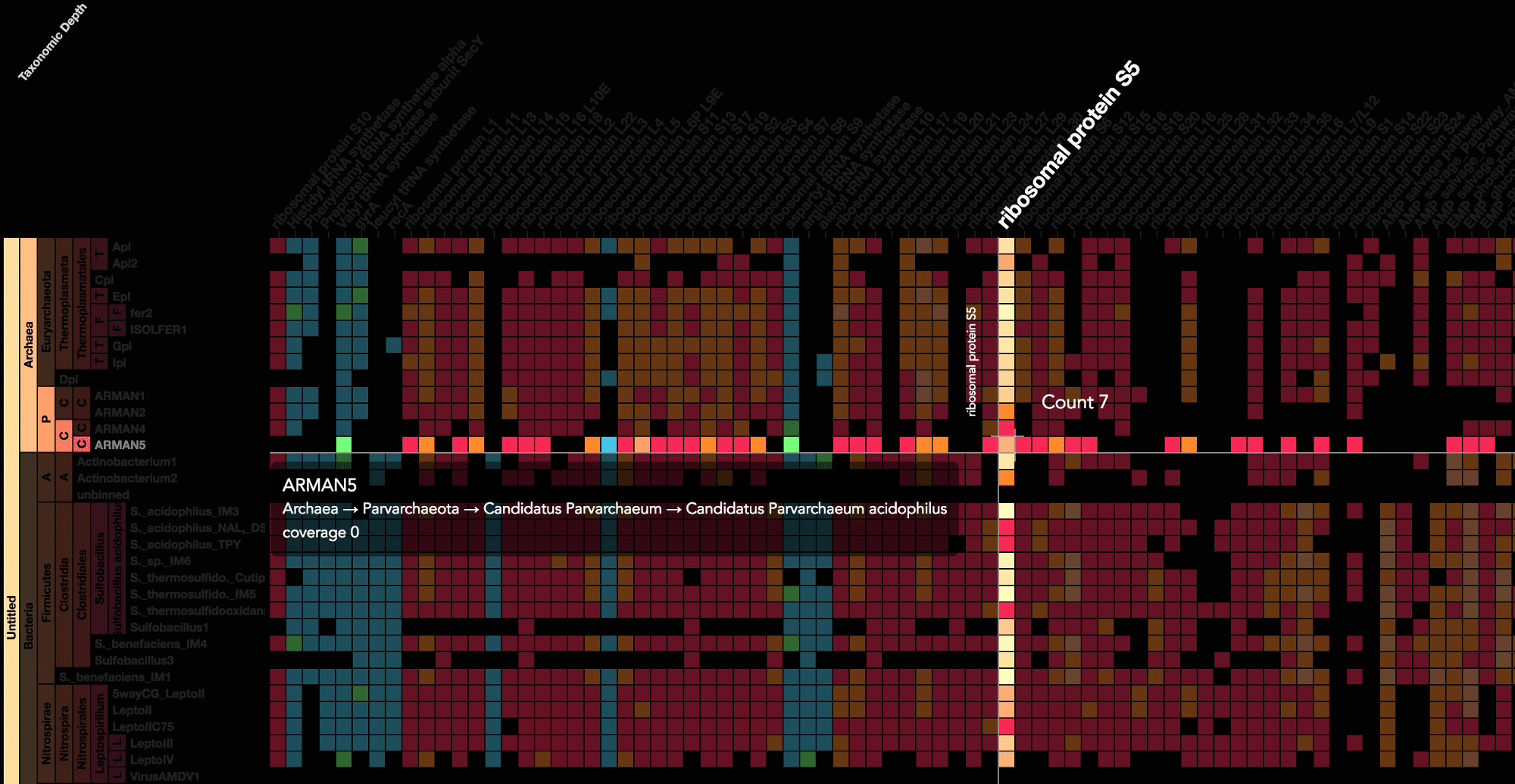
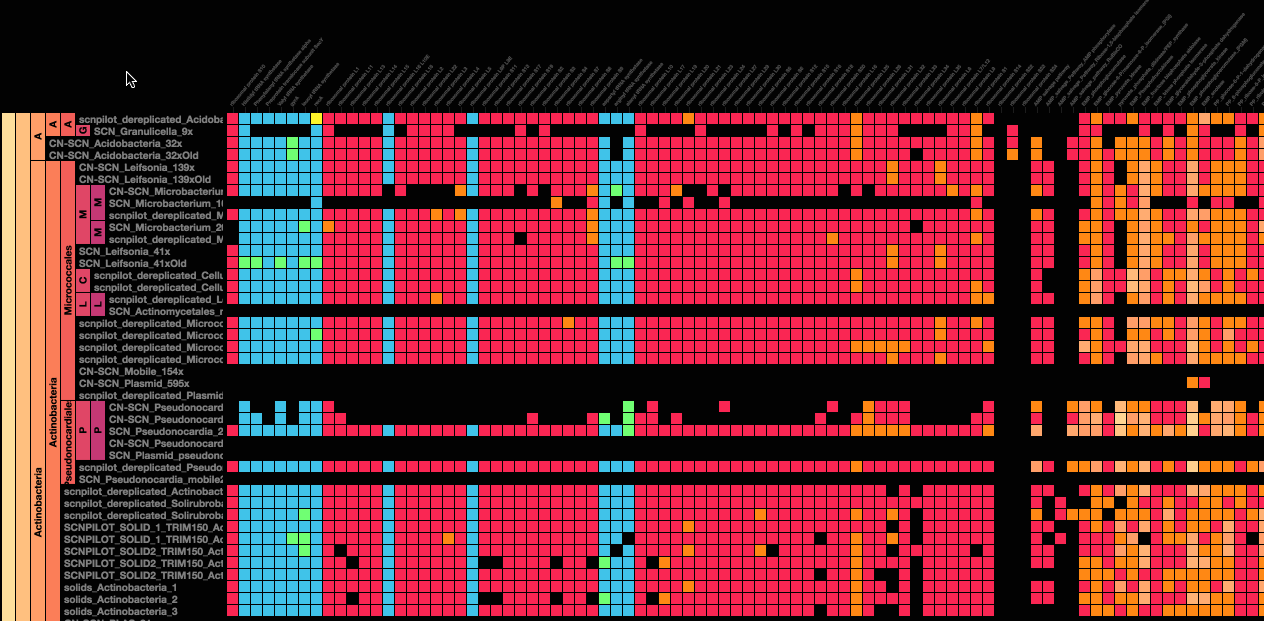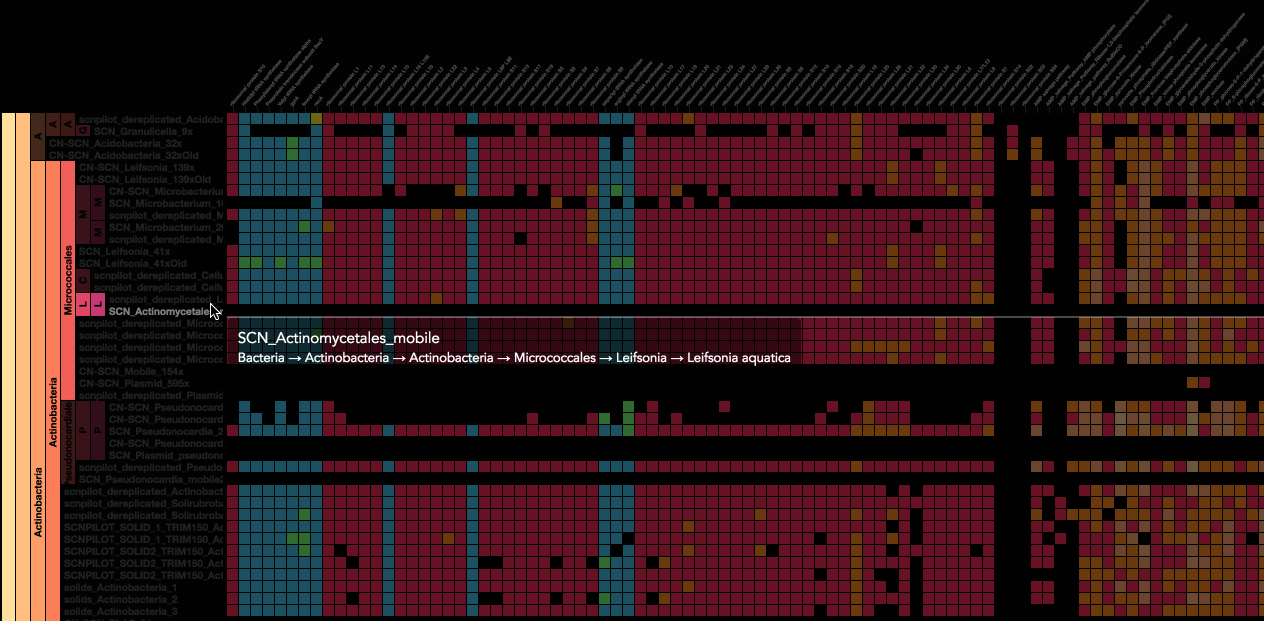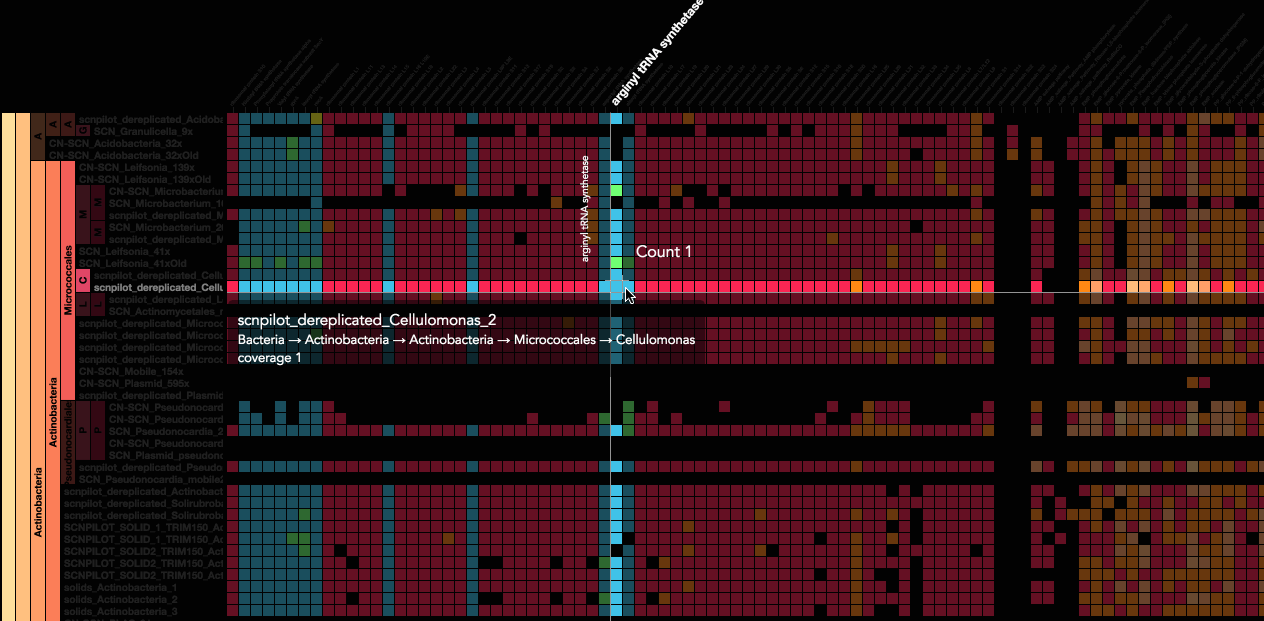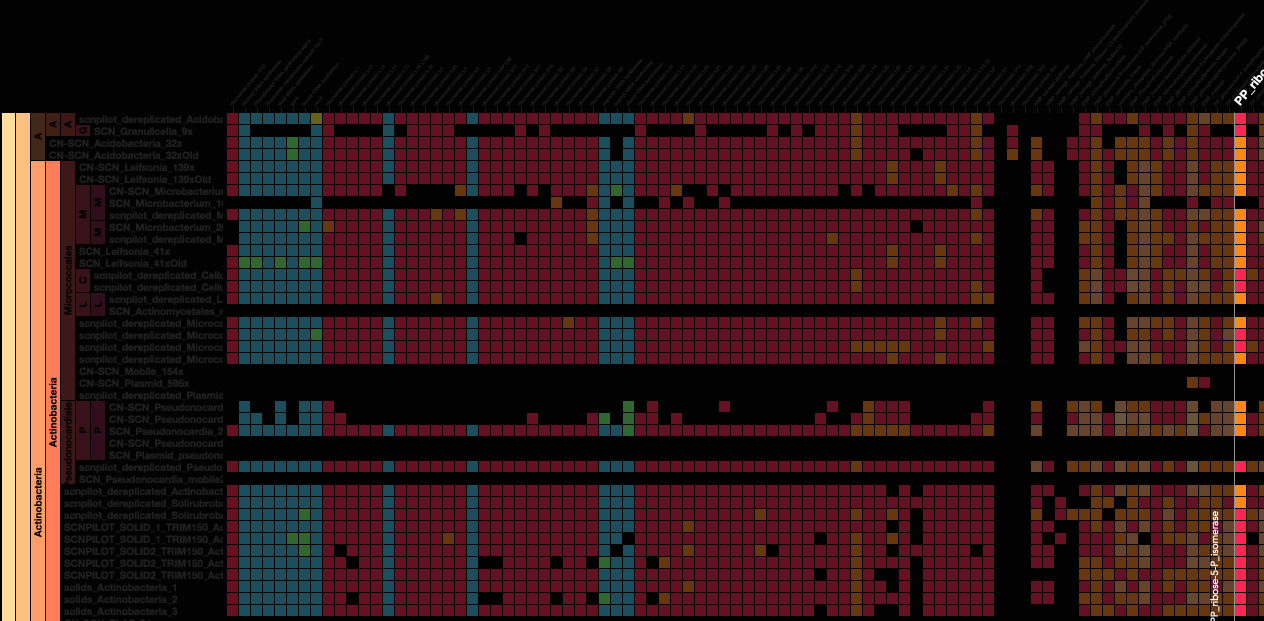
Using advanced data interaction and interactive visualizations, Stamen helped unravel these vast new landscapes of genetic diversity. Visualization is critical to understanding this vast data set. Genome summary visualization displays all the information about a sample in a clear, expressive manner, promoting hypothesis generation and experimentation. Stamen helped Banfield to recreate this genome summary to allow them to visualize even more data simultaneously. Our visualization enabled new variations and interactions with the data, allowing Banfield to ask bigger and more comprehensive questions of these larger samples – for example, to learn which organisms in the environment contribute to carbon or nitrogen flow through the system.
Stamen helped us to recreate the genome summary to allow us to visualize even more data simultaneously. Additionally, these new genome summaries are FAST. This is a common theme in biology (in all science these days really) — there’s more data out there, and investigating it requires adding more and more to your experiments. The enhancements Stamen provided brought new variations and interactions with the data that have help users to query the data in a new ways. Including more data in the visualization allows us to ask bigger/more comprehensive questions of the data. Which organisms in the environment contribute to carbon or nitrogen flow through the system, can now be surveyed from bigger samples. Jill Banfield Director, Banfield Lab, UC Berkeley