It’s easy to think that we’ve discovered most of the species on the planet. In fact, the booming field of metagenomics is using big data to help scientists better understand new and vast unexplored regions of the natural world: the microbiome. In the last few years, the price of genetic sequencing has plummeted to the point where scientists can now study ecosystems in their entirety, not just the parts of it that they already know how to identify.
This new way of working relies on the visualization of truly enormous sets of interrelated data about organisms that often are new to science.
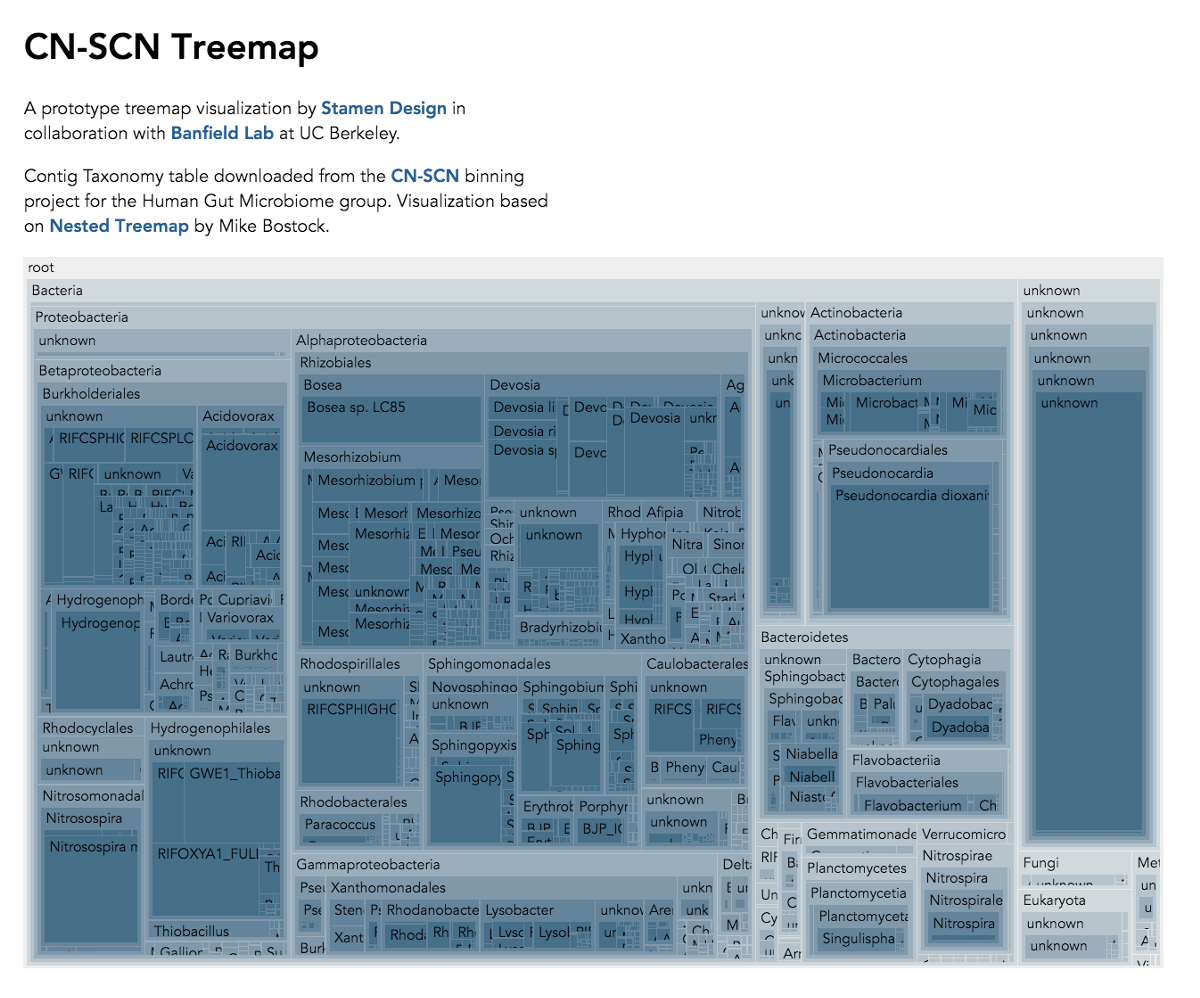
The Banfield Lab at Berkeley recently collaborated with Stamen to bring this data to life, using advanced data interaction and interactive visualizations to make it easier to understand these vast new landscapes of genetic diversity. What follows is an interview I did with Brian Thomas and Jill Banfield at the Banfield Lab about their amazing work at the lab, the work we did together, and where the field is going.
What is metagenomics?
A metagenome is a collection of DNA sequences from the organisms present in an environment at the time a sample is taken. Metagenomics is the study of these genomic sequences. Metagenomics has been around in various forms since the early 2000s. Initially, the approach was referred to as “community genomics” because the sequencing approaches were used to study natural microbial communities. It wasn’t until the mid 2000s that it acquired its “meta” label. In the first studies, both genome reconstruction-based approaches and analyses of collections of genes without organism affiliation were used. Both of these methods are distinct from investigations of specific marker genes that had been used previously to phylogenetically “fingerprint” environments.
The primary difference is that genome sequences provide some insight into what all the organisms might be doing. Fingerprinting methods mostly tell us how closely organisms are related to each other.
The initial metagenomic studies were done on biofilms growing in battery acid—like the acid mine drainage (AMD) streams.
This ecosystem was ideal because most of the DNA sequences came from a few abundant organisms, a feature that simplified the problem of working out which DNA sequences came from which organisms and made it possible to recover genome sequences from organisms outside of the laboratory — for the first time.
Over the next 10 years, this AMD system became the most extensively studied ecosystem ever, with over 80 publications on topics ranging from how AMD organisms accelerate acid formation to the first measurements of evolutionary rates of bacteria in their natural settings. It was the “bedrock” research topic of dozens of PhD students and post-doctoral fellows, the majority of which are now faculty members at several institutions in the USA and other countries (Luxembourg, Sweden, Australia) .
Why is it new? How is it different from “regular” genomics?
Genomics has revolutionized what we know about life, all levels of life, from viruses, to single-celled organisms, to the multicellular life we can actually see. The “problem” with the first-developed form of genomics, however, is you had to know what it is you wanted to sequence and you had to have the organism in a pretty pure form before you could access a clean genome DNA signal.
Most often the organisms available were of interest because they were known to impact human health, i.e., they were mostly pathogens. To a lesser degree we knew about genomes from organisms with economical/agricultural importance to humans. Other than that, the collection of organisms suitable for “regular” genome sequence was pretty minimal.
This is why metagenomics was different, and why it’s amazingly powerful. You can go into ANY environment and you can access essentially ALL the genomes… in an unbiased manner. This is incredible, and as you can guess, where you didn’t think there were any life forms to be seen, you discover amazing new organisms that nobody even knew existed.
The findings drive home how insignificant the “human-related” genomes are, when compared to everything else that’s out there.
Why is it important? Where is it going?
DNA sequencing is getting cheaper and better every year. As the cost of generating DNA sequences continues to go down, it will become increasingly common throughout science.
Everyone will have their genomes sequenced — and they will also profile the microbiome of their homes, workplaces, food and pets. Every environment will be sequenced, multiple times each day or month or year.
From the basic science perspective, Instead of spending time and money figuring out how to get an organism to grow in a lab, you’ll find it in its natural environmental setting and start with its genome. Now you’re armed with an understanding of who the organism is, and what that organism is capable of. From this, you can make predictions about how to get it to grow in the lab, or what it’s doing in the environment or how it’s interacting with its neighbors. Metagenomics is critical for biology and ecology in the future.
What role does visualization play in helping to do this science?
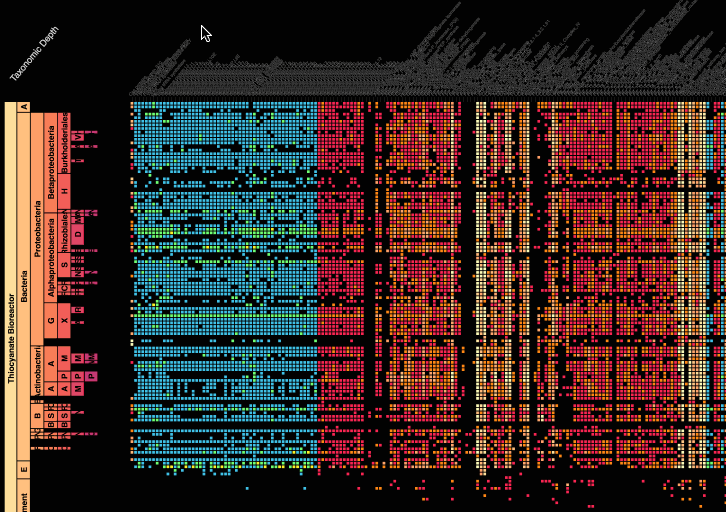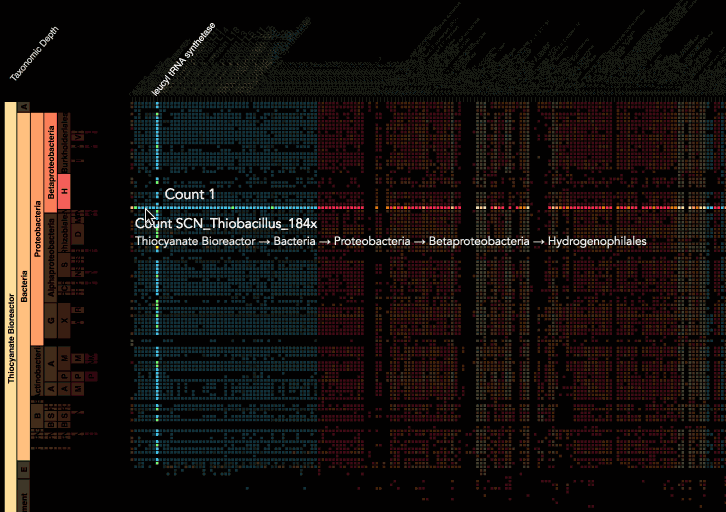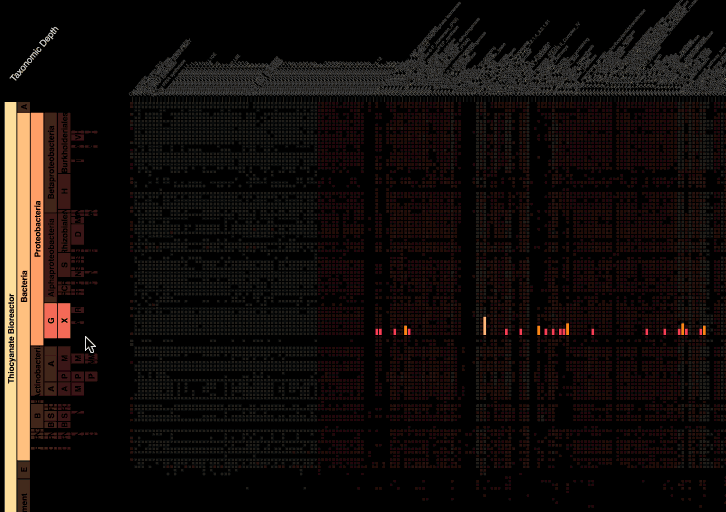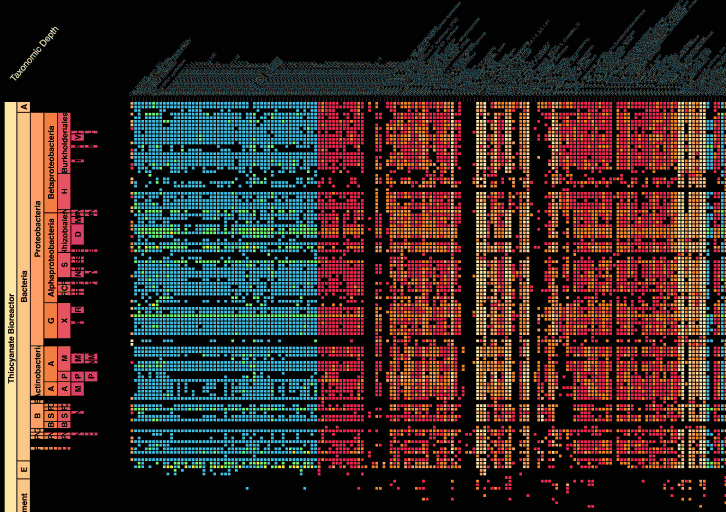
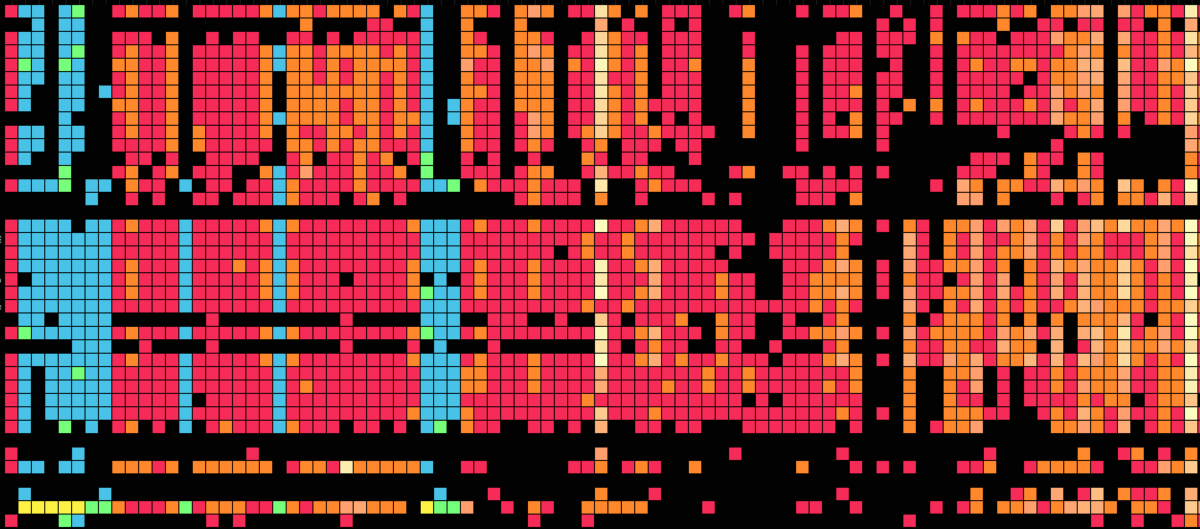
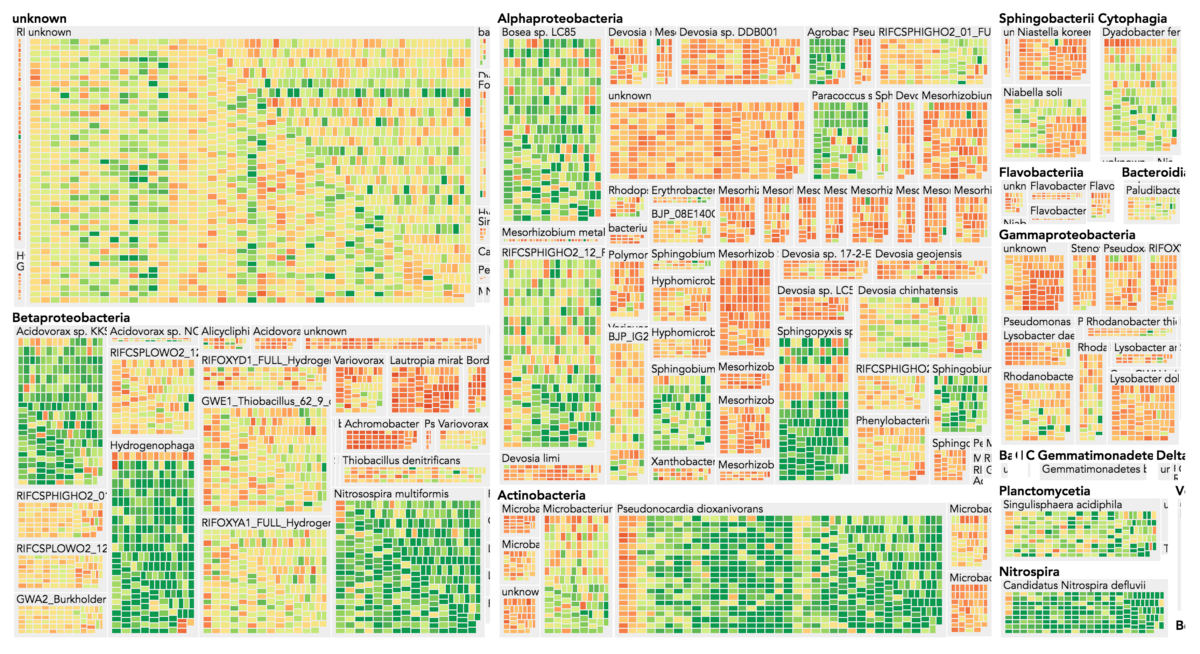
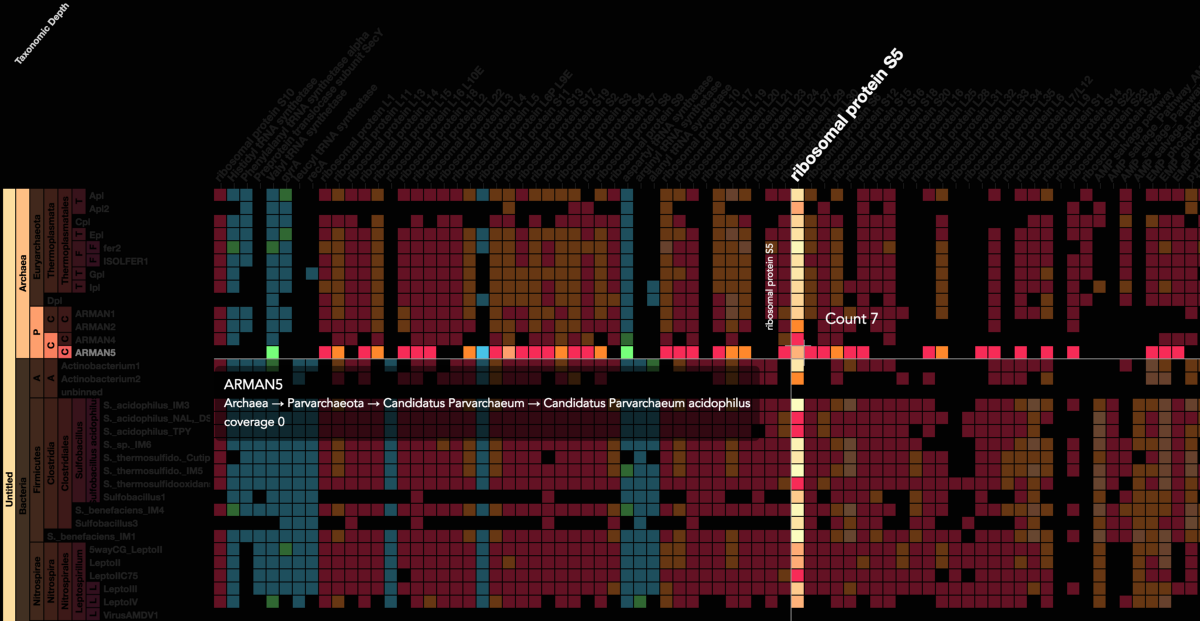
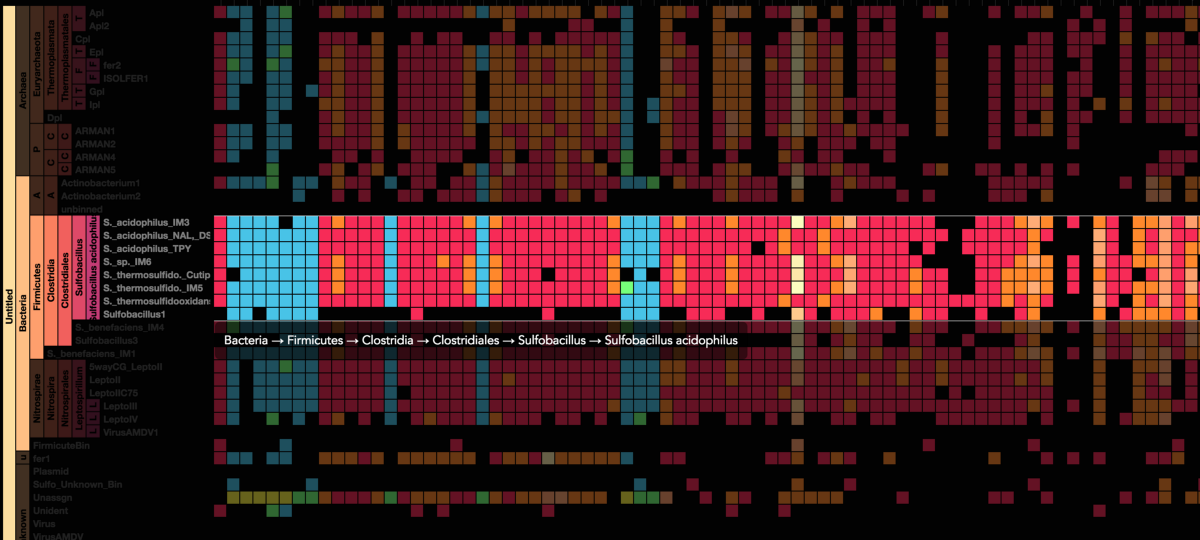
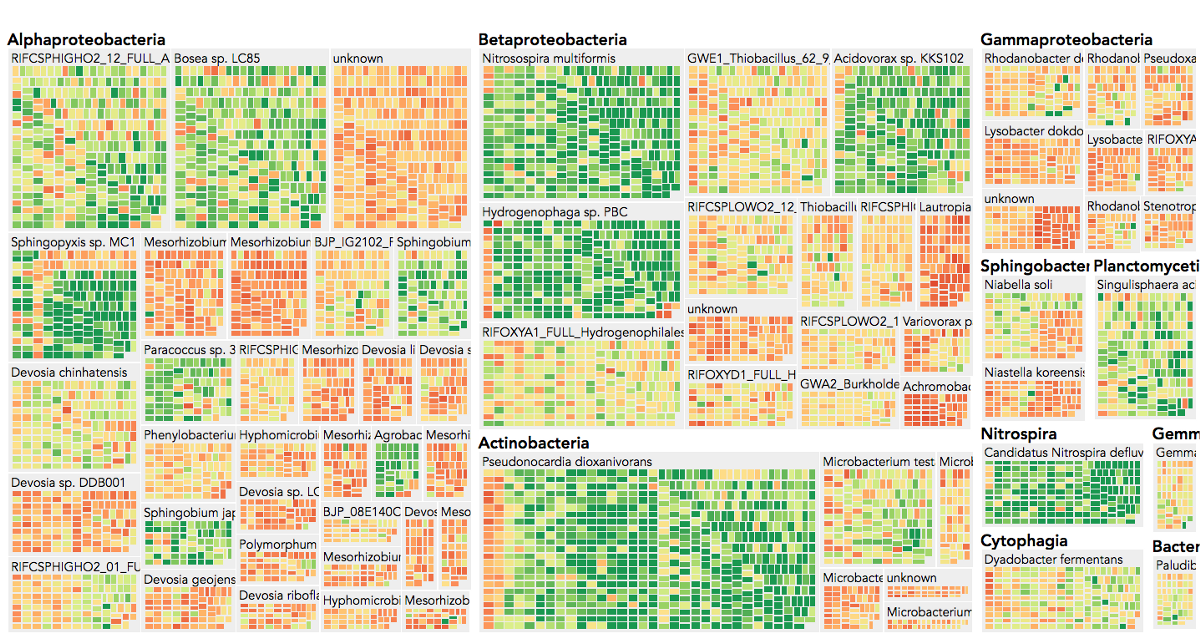
Visualization is critical, especially in metagenomics. You can’t see this life directly (without some pretty powerful microscope technology, and even then you don’t know who you’re looking at!). Having metagenomes from an environment however, allows us to understand what organisms are doing and have the potential to do. We use a visualization called a “genome summary” extensively.
This visualization displays all the information we’ve collected about a sample in a clear, expressive manner, that promotes hypothesis generation and experimentation. The amount of data that goes into a genome summary display is extensive. The system we’ve developed tracks it all, and this feeds the visualization. Metagenomics is definitely big data, and the genome summaries allow us to look at all the recovered organisms and all metabolic reactions of these organisms simultaneously.
How is the work that Stamen built for you helping you do your work?
Stamen helped us to recreate the genome summary to allow us to visualize even more data simultaneously. Additionally, these new genome summaries are FAST. This is a common theme in biology (in all science these days really) — there’s more data out there, and investigating it requires adding more and more to your experiments. The enhancements Stamen provided brought new variations and interactions with the data that have help users to query the data in a new ways. Including more data in the visualization allows us to ask bigger/more comprehensive questions of the data. Which organisms in the environment contribute to carbon or nitrogen flow through the system, can now be surveyed from bigger samples.
How did you get involved?
A very long time ago (~1984) I became fascinated by the possibility that microorganisms impact processes that are ongoing around us at the Earth’s surface — in soils, water and sediments.That wasn’t something we talked about much in my undergraduate training. In the mid-1990s I decided that it was time for me to study this, and the obvious case involved bacteria that were known to directly accelerate sulfide mineral dissolution and so had direct environmental impact. By that time in biology it was becoming obvious that the few organisms that had been studied in the lab probably did not represent the full diversity of organisms “out there”.
Our first “baby steps” were to work out which organisms were actually present at sites where AMD was forming by fingerprinting the marker genes of the organisms present. But knowing who the organisms were didn’t tell us how they were involved in the process. By the later 1990’s genomes for laboratory-accessible organisms were becoming available for the first time. Thus, our first step was to sequence the genome of an organism that had been isolated from our AMD system. This was interesting, but it raised the question: “What about the rest of the organisms out there — those that we don’t yet have growing in the lab”?
So, we proposed to sequence all of the organisms simultaneously — with some hope that we could sort it out after the fact (no clear plan). Remarkably, the US Department of Energy (program manager: Dan Drell) decided to support this idea. We waited in line for our AMD biofilms to get onto the sequencers — in the queue behind the human genome project. Eventually this happened, and pretty much as soon as we had the data, it was clear that the approach worked.
In very early 2004, we published the first ever genome sequences for organisms that were not growing in a lab.
It took quite a few more years before sequencing became cheap enough and available enough to make it possible to scale up our methods to study really complex environments such as soils and aquifer sediments. But we did.
Of the many discoveries you’ve made, which were you the most amazed/shocked/confounded by?
That is easy to answer! We’d been working on AMD for quite a few years — as one of the only groups that were studying genomes of organisms directly sampled from the environment. We got involved in a project that was studying a uranium-impacted aquifer and the microbiology of the system became a critical question.
Thus, we transferred our “AMD methods” to this complex system expecting to just learn some basic information about variety within the set of laboratory-studied organisms that were believed to be important there. When we analyzed our first metagenomic sequence datasets we were ‘blown away’. First, most of the genomes looked nothing like anything that had been seen before. Ultimately we linked many of the genomes to organisms only known through cultivation-independent gene surveys. Second, and particularly striking was the fact that some of the organisms used a different genetic code.
Finally, it was apparent that the genomes that we sampled were tiny, novel, and probably from organisms that could only grow as part of microbial communities because the organisms seem to lack the ability to do many of the things needed for “life”. Some of the organism groups detected in that first study we now recognize as completely new lineages on the tree of life.
A few months ago we published data that suggests that this previously essentially invisible, enigmatic group may exceed — in terms of diversity — all other bacteria. I doubt that anything like the full implications of these findings have been recognized yet in the community of biological scientists.
What’s the difference between the inside of a killer whale’s mouth and a dolphin’s mouth? Why is this important?
We know they are uniquely different. Natasha Dudek and David Relman from Stanford are using our methods to investigate microbial samples from sea mammal mouths. It’s very interesting to see that in a habitat you’d think is pretty similar (two sea mammals, both eating the same food source) you find very distinct collections of microorganisms. This difference really accentuates the fact that we know very little about our (multicellular life’s) microbiomes.
Some estimates recently have shown that there are the same number of bacteria in and on the human body as there are human cells in the body.
This is undoubtedly the case across the spectrum of complex life forms on the planet (possibly even a clue to their complexity). And we really know very little about this “unseen” life — the microbiome. As mentioned, if it doesn’t cause us a problem, we probably don’t know about it. It really begs the question, “what is normal?” We know that disease results from abnormalities, an ecosystem getting out of balance for example, like an infection. But what is normal? Metagenomics is really the key to understanding this.
Some estimates recently have shown that there are the same number of bacteria in and on the human body as there are human cells in the body. This is undoubtedly the case across the spectrum of complex life forms on the planet (possibly even a clue to their complexity).
There will be many advances that directly stem from understanding microbiomes through metagenomics. New drugs, antibiotics, industrial chemistry advances etc. More importantly, as we start to get a true understanding of the unseen life around us and on/in us, we’ll be able to leverage that information.
Imagine the knowledge that would allow us to influence an ecosystem to achieve a particular outcome. Instead of prescribing a drug to kill an overgrowth of a “bad bug” on a human, we can promote the growth of the other “good bugs” on that person to outcompete the invader. Or imagine the next time there’s an oil spill in an ocean. We’ve seen how the normal, background bacteria in the ocean that are at low abundance rise up to eat the new food source (oil), and lessening the impact of the disaster for us and the environment. As we start to know more about the full extent of microbial life in these environments, you could imagine “stimulating” the recovery process after an accident by promoting the growth of these background players.
How are you leveraging the 3 billion years of evolution that microbes/viruses enjoy — to do what ?
Understanding the genomic content of an ecosystem yields incredible insight into who the dominant organisms are, the minor constituents, and all levels in between… including viruses. You can see the changes that happen within and between organisms, how genetic information is shared, and how predation can alter community structure. Microbes are presumed to be the first life forms that appeared on the planet: viruses, and the organisms present today are the result of the evolutionary forces in these ecosystems.
The insight we gain from understanding an organism from its genome sometimes yields some unexpected results, like the CRISPR-Cas system in Bacteria. This is a viral defense mechanism that helps a bacteria to evade predation by viruses.
What’s neat about this system is that it chops up the viral genome into little bits and then actually splices little bits back into its own genome. Then, the next time this bacteria (or its progeny) are exposed to this virus, it has the blueprint for the virus and thus will be immune.
Of course, the virus fights back! It mutates its own genome to try to overcome its host’s immunity. So it’s like an arms race between the two. When you look that the viral incorporation signature on the genome itself, it’s arranged in a unique way that allows us to trace viral predation events through time. This system captivated a generation of biochemists who, protein-by-protein deciphered how it works.
Now CRISPR-Cas is a household name and there are expectations that it will be harnessed to change the world, including by rewriting human DNA to obliterate hereditary diseases. The CRISPR-Cas system is just the tip of iceberg. As we study more genomes from the environment, we’ll find more and more interesting systems like this and the potential value to address societal needs is unimaginable!
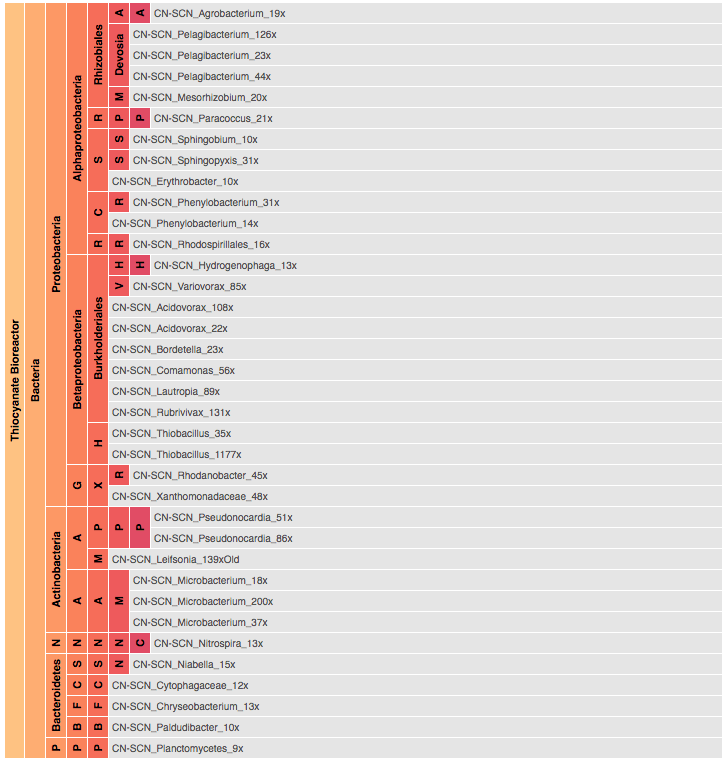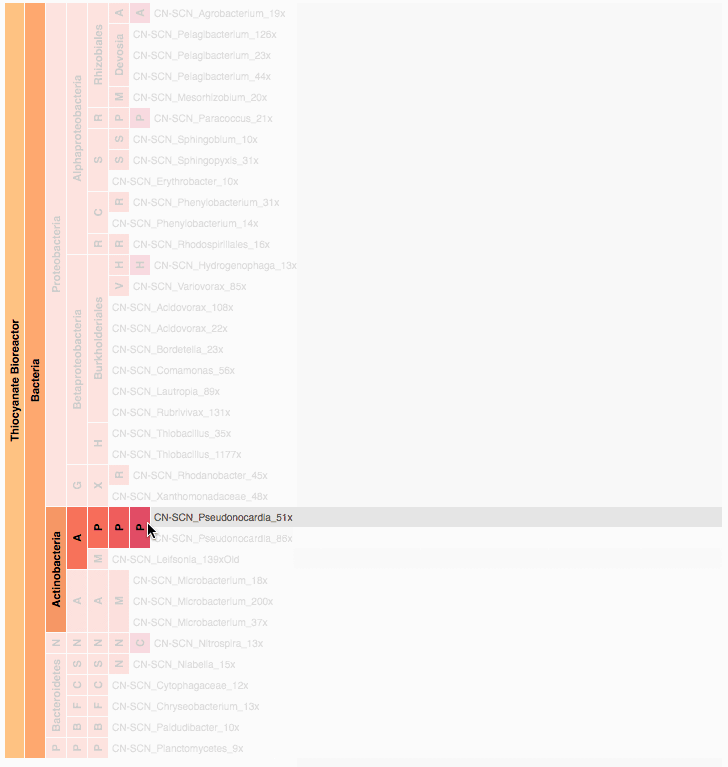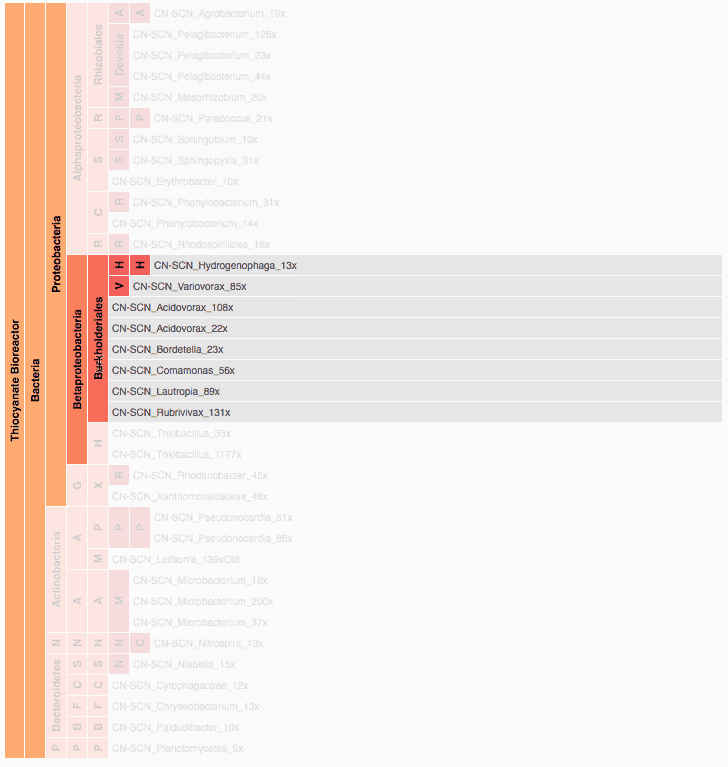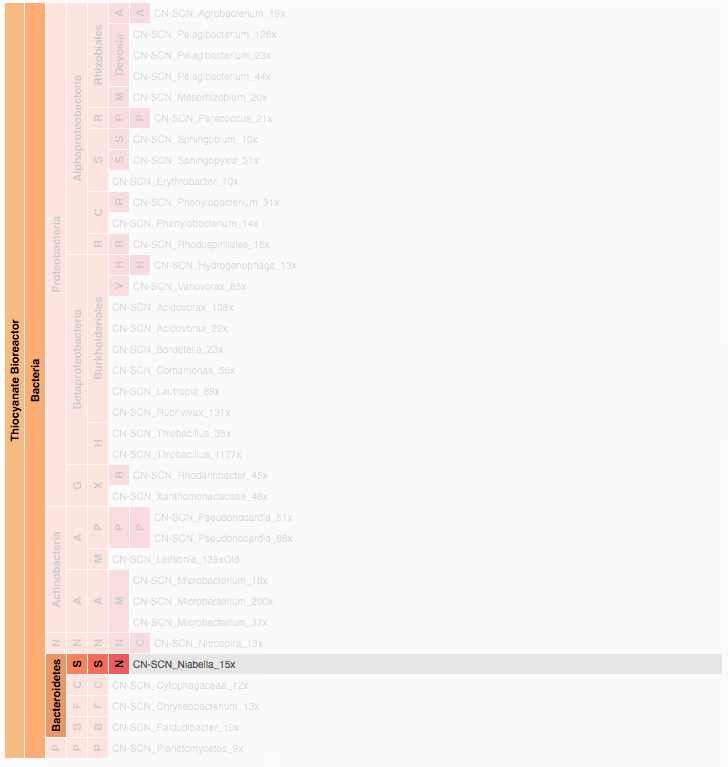
In terms of the future, where is this work going to take us and be applied in public and scientific domains? What’s your vision for what types of breakthroughs metagenomics discoveries will enable? What are some commercial applications? What are some scientific applications? Help paint the picture of how important this work is.
More than ever before, understanding our environment is critical. As the planet heats up, environments are going to change. As the population continues to increase beyond sustainable limits, things are going to change. Humans worry about what they can see, but it’s the microbes on the planet that are critical to every geochemical cycle. Metagenomics will give us the data signal we need to develop better tools to monitor our environment… REALLY monitor it, all the way down to every species.
Metagenomics will be our savior as a reservoir for new medicines. Take the recent discovery of a new antibiotic, isolated from our own normal nose microbiome! We’re literally walking around with a community of microorganisms on and in us that potentially can answer many of our modern day health-related issues.
Tapping this knowledge will be invaluable. We feel that being able to accurately monitor environments is critical, and we think the tools that we have begun developing are just the beginning. One of the areas we’re planning on focusing is contaminated environment monitoring. Contaminated environments (e.g. from mining or other industrial processes etc) are huge global health concerns. We think metagenomics is going to play a key role in tackling these problems.
To read more about the amazing work that Banfield’s lab is going, take a look here, and to find out more about how Stamen is working with scientists to do their work, visit https://stamen.com/science/.
